現代作物在產量與品質上表現優異,但其背後卻潛藏著遺傳多樣性貧乏的風險。當前的栽培品種,多因長期的人為馴化 (domestication) 與高度篩選,導致遺傳背景變得十分狹窄,在追求產量與商業價值的過程中,許多原本存在於自然界的優異抗性基因被捨棄,導致栽培作物在面對病原菌演化或環境壓力時,缺乏足夠的基因緩衝空間。
相較之下,散佈於自然界中的野生近緣種 (wild relatives) 經歷了長期的天擇與演化,累積了強大的環境適應與病蟲害防禦機制,透過引進這些具利用價值的野生基因,將其導入栽培品種中,已成為提升現代作物抗逆境的關鍵路徑,例如,栽培西瓜經常受到蔓枯病 (Gummy Stem Blight) 的威脅,育種家透過篩選非洲的野生西瓜品系,成功找到了能抵抗病原菌的基因;在小麥育種領域,科學家利用野生的山羊草屬 (Aegilops spp.) 作為抗銹病基因的來源,藉此在農田中建立穩固的抗病防線。
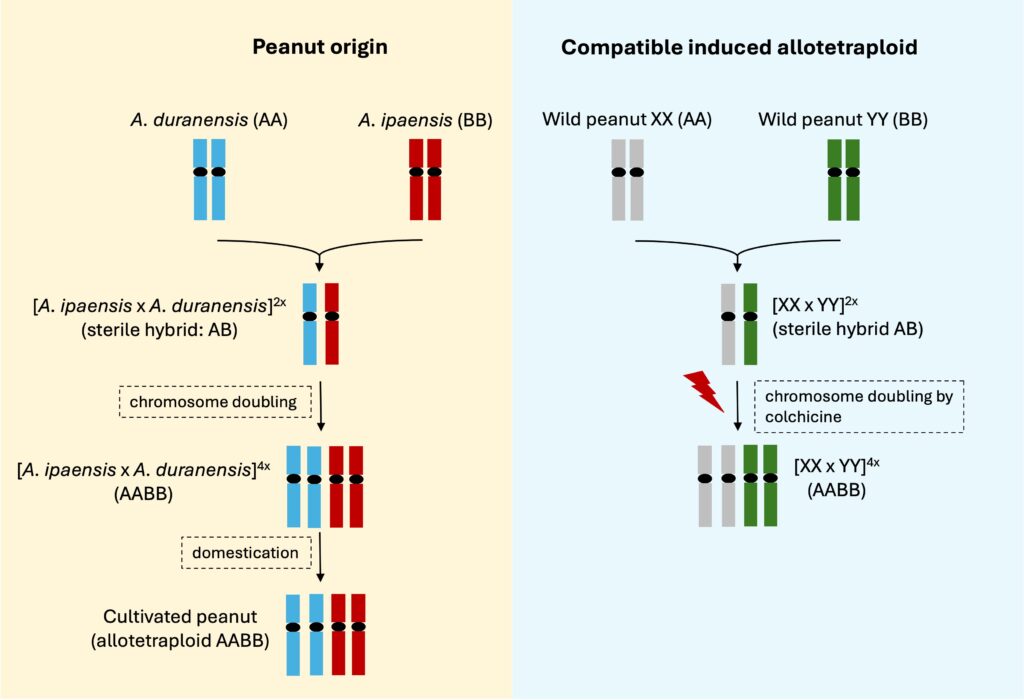
花生起源於南美洲,其演化過程約發生在一萬年前,由攜帶兩種基因組的野生二倍體花生(Arachis duranensis – 基因型為AA與Arachis ipaënsis – 基因型為BB),因人為攜帶導致生長地域重疊,在自然界中發生了跨物種雜交,該雜交產生了一個不育的二倍體 (sterile hybrid, 基因型為 AB),隨後在自然界該二倍體發生染色體倍增 (chromosome doubling),演化成了擁有四套染色體 (4n, 基因型為 AABB) 的異源四倍體 (allotetraploid) 花生始祖。這意味著現代栽培花生 (cultivated peanut, 基因型 AABB) 同時繼承了兩個野生祖先的遺傳物質。雖然這種演化路徑賦予了花生獨特的生理特性,卻也使其與其他野生二倍體親戚間產生了嚴重的生殖障礙,導致栽培花生的遺傳背景極為狹窄,嚴重限制了可用於作物改良的資源。
在南美洲的玻利維亞、阿根廷及巴西一帶的荒野中,科學家至今已記錄了 80 種以上的野生花生,這些野生種在自然生境中,長期與環境中的真菌、線蟲及害蟲共同演化,基因組中蘊藏著栽培花生所缺乏的抗性多樣性,例如,野生花生 Arachis cardenasii 具有極強的葉斑病及根瘤線蟲抗性,這些寶貴的資源已成為全球花生遺傳改良的重要基石,以美國商業品種 Tifguard 為例,它正是導入 Arachis cardenasii 的抗性基因片段,獲得了卓越的根瘤線蟲抗性,並在田間表現中展現出顯著的產量提升與抗病成果。然而,將野生花生基因導入栽培品種存在天然的生殖障礙,由於栽培花生屬於異源四倍體,而多數具備強大抗性的野生花生則是二倍體,這種染色體倍性的差異,使得兩者雜交後的後代染色體無法正常配對,導致不育。為了跨越障礙,育種學家運用染色體工程技術模擬現代花生的自然產生過程,首先選取兩種具備不同基因組的野生二倍體花生進行人工雜交,再利用秋水仙素 (colchicine) 處理以誘導其染色體倍增,創造出合成異源四倍體 (induced allotetraploid) ,這個過程成功克服了野生花生與栽培花生染色體倍性不相容的問題,讓新合成的異源四倍體能與現代栽培花生進行有效的遺傳交換,目前這套技術已成熟應用於品種改良,例如巴西品種BRS 425,即是透過此技術導入野生基因,顯著提升其對葉斑病的抗性。
利用野生資源改良作物,核心意義在於強化農業生產的穩定與韌性,這些存在於荒野的資源,透過傳統育種方法或現代精準育種策略重回農田,使作物具備更強的天然免疫力,進而降低糧食生產對化學藥劑的依賴,這不僅提升了作物的耐受力,也讓我們在面對未來氣候與環境挑戰時,擁有更豐富的遺傳籌碼。
While modern cultigens prioritize crop yield and quality, they harbor a hidden risk: a profound lack of genetic diversity. Most contemporary cultivars possess a narrow genetic base resulting from long-term domestication and intensive selection. In the pursuit of high productivity and commercial value, many superior resistance genes originally present in nature were inadvertently discarded. Consequently, when faced with evolving pathogens or environmental stresses, cultivated crops often lack sufficient genetic buffering capacity.
In contrast, wild relatives of crops that survived in nature have undergone years of selection and evolution, accumulating mechanisms for environmental adaptation and pest resistance. Introgressing these valuable wild genes into cultivated varieties has become a critical pathway for enhancing crop resilience. For instance, cultivated watermelons are threatened by Gummy Stem Blight; however, by screening wild watermelon lines found in Africa, breeders successfully identified genomic segments that confer resistance to the pathogen. Similarly, in wheat breeding, scientists utilize Aegilops spp., wild relatives of wheat, as a source of rust resistance genes to secure stable crop yields.
The evolution of the cultivated peanut originated in South America approximately 10,000 years ago. Two wild diploid species with distinct genomes, Arachis duranensis (AA genotype) and Arachis ipaensis (BB genotype), were introduced into the same geographical region because of human activities. This resulted in a natural interspecific hybridization, producing a sterile diploid hybrid (AB genotype). Subsequently, a spontaneous event of chromosome doubling allowed this hybrid to evolve into the ancestral allotetraploid peanut. Consequently, modern cultivated peanuts (AABB genotype) inherited genetic material from two distinct wild ancestors simultaneously. While this evolutionary path endowed peanuts with specific physiological traits, it also established a reproductive barrier against their wild diploid relatives. This isolation led to a narrow genetic base in peanut, significantly restricting the resources available for contemporary crop improvement.
In the regions of Bolivia, Argentina, and Brazil, scientists have documented over 80 wild species in the genus Arachis. These species have co-evolved with fungi, nematodes, and pests in their natural habitats, harboring primitive resistance genes that are absent in cultivated peanut varieties. These South American wild resources serve as an essential genetic reservoir for global peanut improvement. For example, the wild species A. cardenasii possesses resistance to leaf spots and root-knot nematodes. A success story is the U.S. commercial cultivar ‘Tifguard’, which obtained root-knot nematode resistance by introgressing genomic segments from A. cardenasii, significantly enhancing its field performance. However, introgressing genes from wild peanuts into cultivated varieties faces the ploidy barrier. Since cultivated peanuts are allotetraploid while most resistant wild species are diploid, this discrepancy in ploidy levels prevents proper chromosome pairing in hybrids, leading to sterile progeny. To overcome this, breeders employ chromosome engineering to simulate the peanut’s natural evolutionary process: crossing two wild diploid species and treating the resulting hybrid with colchicine to induce chromosome doubling. The creation of these induced allotetraploids successfully overcomes the ploidy incompatibility, allowing the new synthetic allotetraploids to exchange genetic material with modern peanuts. This technology has been successfully applied to resistant cultivar development; for example, the Brazilian cultivar BRS 425 utilized this technique to introgress wild-derived segments, significantly enhancing leaf spot resistance.
The goal of utilizing wild resources to improve crops lies in strengthening the stability and resilience of agricultural production. Whether through traditional breeding methods or modern breeding strategies, integrating these wild genetic assets into our agricultural systems fortifies crops with enhanced resistance. This approach reduces our reliance on chemical applications and provides a more robust genetic toolkit to navigate future climatic and environmental challenges.
參考文獻
Krapovickas, A., et al. (2007). Taxonomy of the genus Arachis (Leguminosae). Bonplandia, 16, 7-205.
Holbrook, C. C., & Stalker, H. T. (2003). Peanut breeding and genetic resources. Plant breeding reviews, 22, 297-356.
Leal-Bertioli, S. C., et al. (2015). Arachis batizocoi: a study of its relationship to cultivated peanut (A. hypogaea) and its potential for introgression of wild genes into the peanut crop using induced allotetraploids. Annals of Botany, 115(2), 237-249.
Suassuna, T. D. M. F., et al. (2019). BRS 425: the first runner peanut cultivar related to wild ancestral species. Crop Breeding and Applied Biotechnology, 19, 373-377.
作者簡介

專長為真菌學、傳統及分子輔助抗病育種、族群遺傳學、基因體學(基因體組裝與註解、生物資訊)、植物病原流行病學、化學藥劑功效評估

