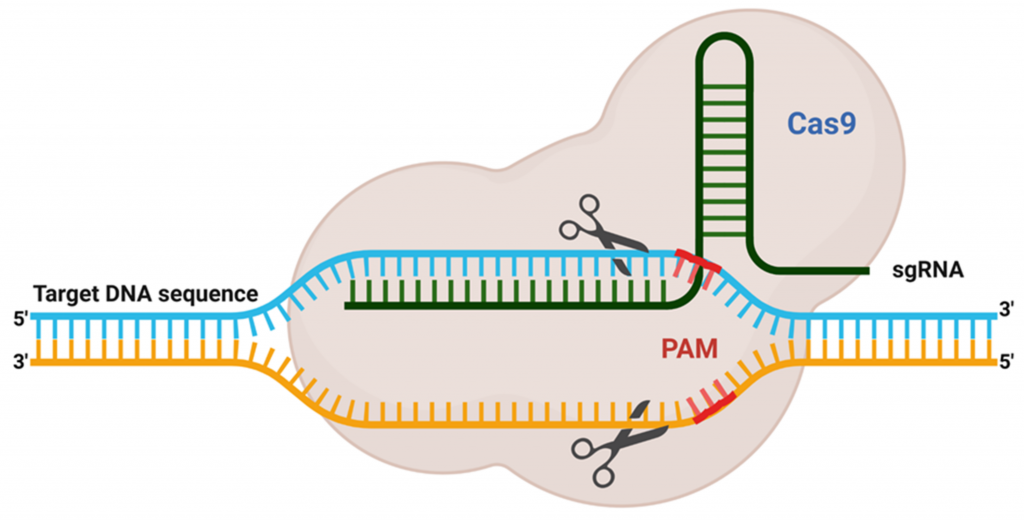
自2012 年Cas9 如何針對目標序列進行編輯的機制被解開後,CRISPR/Cas9 開啟了基因編輯革命性的進展(Jinek et al., 2012; Wiedenheft et al., 2012)。CRISPR/Cas9 為細菌與古細菌的自我防禦系統,當細菌受到噬菌體、病毒或外源質粒攻擊後,啟動自我防禦系統以分解入侵外源 DNA,並且會留下一段外來的 DNA 片段於細菌基因體中,形成重複的特殊序列稱 CRISPR(Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005)。當再次受到相同的病毒或外源 DNA 攻擊時,細菌會轉錄出 CRISPR RNA(crRNA)、transactivating crRNA(tracrRNA)和引導核糖核酸(guide RNA, gRNA),當中Protospacer 序列(20~24 nt)為辨認出病毒或外源 DNA,而在 Protospacer 序列後的三個核苷酸(NGG)稱為 Protospacer adjacent motif(PAM) (圖一),Cas9 核酸酶會辨識 PAM 並切割病毒或將外源 DNA 剪斷,避免再次被感染,觸發基因組 DNA 中雙鏈斷裂(double- strand break, DSB),因此也稱為細菌的適應性免疫系統(Deveau et al., 2010; Mali et al., 2013)。
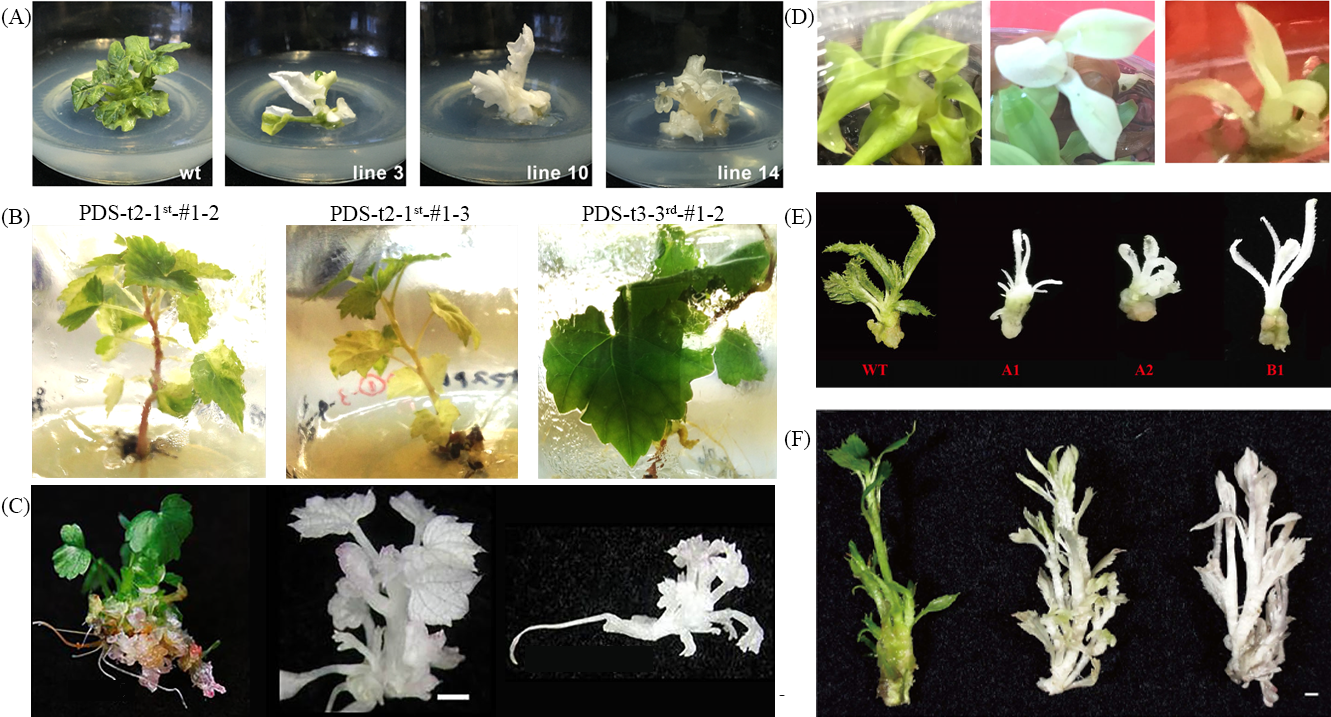
CRISPR/Cas9系統目前為最廣泛運用的基因編輯技術。包括用於治療人類疾病,如癌症、遺傳疾病、病毒性疾病,以及用於動植物、細菌和真菌基因編輯。此外,次世代核酸定序技術 (Next Generation Sequencing, NGS)的快速發展,並且全基因組解序的價格越來越便宜,並可以快速且完整的解序 (Levy and Myers, 2016),在開花植物之基因組定序並且註解超過270種以上的植物 (參考Phytozome v13) (Goodstein et al., 2012; Chen et al., 2019; Marks et al., 2021)。提供了許多植物基因組的序列,從中分析有興趣或是對於植物抗生物及非生物逆境、高產量和抗病相關基因,之後透過基因編輯技術改變該植物的性狀,加速育種速度。該技術目前為止已成功應用在多種植物上,如阿拉伯芥(Feng et al., 2014; Li et al., 2013)、煙草(Li et al., 2013)、水稻(Shan et al., 2013)、小麥(Zhang et al., 2016)、香蕉(Tripathi et al., 2019)、奇異果(Wang et al., 2018)和草莓(Feng et al., 2018)。至於 CRISPR/Cas9 技術應用以糧食作物最為廣泛使用,而在園藝作物上的應用相對較少,主要是園藝作物品種多樣性、建立植物組織培養和基因轉殖系統需要花較長時間,以及園藝作物遺傳背景相當複雜,其中大多為多倍體化、基因體序列資訊不全,因此 CRISPR/Cas9 廣泛應用於園藝產業上仍有其困難度。利用 CRISPR/Cas9技術於園藝作物進行基因編輯,可以用來改變花色、香氣、花型、開花時間、抗病、延緩老化和耐生物及非生物逆境等。而園藝作物以 CRISPR/Cas9 技術進行基因編輯與改變植物色素相關的研究,如 Klimek-Chodacka et al.(2018)編輯胡蘿蔔中的flavanone-3-hydroxylase(F3H)基因,造成原本累積花青素的癒傷組織轉變成白色。而關於改變類胡蘿蔔素是目前以 CRISPR/Cas9 技術針對園藝作物進行基因編輯發表最多之研究,在這些研究中都以 phytoene desaturase(PDS)基因為目標基因進行編輯,當 PDS 基因被剔除後,所有植物都表現出白化性狀(albino),如西瓜(Tian et al., 2017)、葡萄(Nakajima et al., 2017)、草莓(Wilson et al., 2019)、香蕉(Kaur et al., 2018)、奇異果(Wang et al., 2018)和蘋果(Nishitani et al., 2016)(圖二)等。
(一) 提高作物質量和改變農園藝性狀
花卉作物基因編輯相關研究,Watanabe et al.(2017)將牽牛花(Ipomoea nil)花青素生合成基因 dihydroflavonol-4-reductase-B (DFR-B)藉由 CRISPR/Cas9 剔除後,得到白色及粉紅花色的牽牛花;而相同策略在夏堇中,則針對 F3H 基因進行編輯,也獲得白、粉紅及淡紫花色的夏堇(Nishihara et al., 2018)(圖三)。Nonaka et al. (2017) 將番茄中的Glutamic acid decarboxylase 基因 (GAD2、GAD3)以CRISPR/Cas9方法造成突變,可以提高番茄果實中的gamma-aminobutyric acid (GABA)含量(圖四)。另外,剔除掉GABA transaminase (GABA-TP1/2/3)和 Succinate Semialdehyde dehydrogenase (SSADH)基因也可以提高番茄果實GABA含量 (Li et al., 2018)。酵素性褐變 (enzymatic browning)在蔬果中發生在受到壓擦傷、加工、及貯藏,能夠引起蔬果外觀的劣變,導致蔬果的風味和營養產生變化。多酚氧化酶 (polyphenol oxidases, PPOs)為引起蔬果酵素性褐變酵素。PPO主要是將酚類 (phenol)氧化成二酚類 (diphenol),之後轉變成quinones,為一種不溶於水的褐色聚合物,在植物中造成褐化最主要原因 (Gerdemann et al., 2002),將馬鈴薯中的StPPO2 (González et al., 2020)與茄子中的SmelPPO4、SmelPPO5和SmelPPO6三個同源基因 (Maioli et al., 2020),以CRISPR/Cas9方法剔除,可以讓馬鈴薯減少73%及茄子48~61%酵素性褐變。至於番茄果實顏色的改變,Li et al. (2018) 以CRISPR/Cas9多重基因編輯系統 (CRISPR/Cas9 multiplex genome editing system)將與生合成茄紅素 (lycopene)相關基因括STAY-GREEN1 (SGR1)、lycopene ε-cyclase (LCY-E) beta-lycopene cyclase (Blc)、lycopene β-cyclase1 (LCY-B1)和LCY-B2同時進行基因編輯造成突變,使得番茄果實中茄紅素提高到5.1倍的含量。
(二) 增加植物之抗病性
增加植物抗病的能力一直是園藝作物育種中重要的性狀,柑橘潰瘍病是由柑橘黃單胞菌(Xanthomonas citri subsp. citri, Xcc)所引起,研究發現對於易感染 Xcc 柑橘中,LATERAL ORGAN BOUNDARIES 1(CsLOB1)基因表現量都高於不易感病的柑橘(Hu et al., 2014),因此 Peng et al.(2017)針對 CsLOB1 基因啟動子進行編輯,弱化 CsLOB1 基因表現量,使得柑橘對於 Xcc 具有抗性。另一方面,透過CRISPR/Cas9方法將作物中對於真菌性病原菌易感病基因,進行編輯使得作物具有抗或耐真菌性病原菌。將葡萄(Vitis vinifera)中的VvMLO3 和 VvMLO4 (mildew resistance Locus O)同時編輯造成突變,可以讓葡萄對葡萄白粉菌(Erysiphe necator)具有抗性 (Wan et al., 2020)。而關於應用CRISPR/Cas9方法使作物對於病毒具有抗性有以下幾種策略。針對番茄黃化捲葉病毒 (Tomato yellow leaf curl virus,TYLCV)設計會與TYLCV 中的coat protein (CP)或 replicase (Rep)序列互補之sgRNA,而透過基因轉殖,番茄中會形成Cas9-sgRNA complex會針對TYLCV的CP及Rep序列進行切割,造成TYLCV無法在番茄中進行複製進而降低其病毒量 (Tashkandi et al., 2018)。植物中的eukaryotic initiation factor 4E (eIF4E)和eIFiso4E (Lellis et al., 2002; Nicaise et al., 2003; Ruffel et al., 2006)為植物RNA病毒在感染植物時所必須的因子。因為eIF4E和eIFiso4E會和植物RNA病毒中的viral-encoded protein VPg 結合,共同協同病毒複製 (Duprat et al., 2002)。研究指出eIF4E和eIFiso4E對於植物上是多餘的既使他發生突變對於植物的生長沒有任何的影響 (Duprat et al., 2002; Jackson et al., 2010; Wang and Krishnaswamy, 2012; Sanfaçon, 2015)。因此在植物對於potyvirus具抗性是因為eIF4E和eIFiso4E發生突變而獲得對於病毒具有抗性 (Gómez et al., 2009)。
(三) 改變植物二次代謝物
自古以來,人類就使用藥用植物作為食物、治療和工業用途。它和糧食作物一樣在人類文明和社會發展過程中,佔極重要的角色 (Dey et al., 2021)。現代研究人員利用生物技術干預一些重要藥用植物中次級代謝物生合成途徑,以提高或改變其有效成份,可用於醫療、飲食和工業用途 (Dey et al., 2017)。由於NGS發展及基因編輯技術成熟,針對藥用植物進行修改次級代謝物途徑變的容易。
罌粟 (Papaver somniferum)是古老藥用植物,作為多種疾病的止痛,為多種鎮痛劑的來源。罌粟中的嗎啡 (Morphine)、蒂巴因 (Thebaine)、可待因 (Codeine)、罌粟鹼 (Papaverine)和那可汀 (Noscapine)等苄基異喹啉類 (benzylisoquinoline alkaloids, BIAs)都有重要的醫用價值 (Fairbairn and Wassel, 1964; Valva et al., 1985)。在罌粟中benzylisoquinoline alkaloids生合成路徑中的3’-hydroxy-N-methylcoclaurine 4’ -O-methyltransferase (4’OMT)其最終產物為嗎啡、蒂巴因、罌粟鹼、可待因和那可汀 (De-Eknamkul and Zenk, 1992; Morishige et al., 2000)。Alagoz et al. (2016)以CRISPR/Cas9 方法將4’omt2 基因剔除,在4’omt2 罌粟突變株中,牛心果鹼 (S-reticuline)的生合成量是下降最多,而由它所生合成出的嗎啡、那可汀、蒂巴因、罌粟鹼和可待因顯著的下降,甚至不合成此類物質。因此透過基因編輯方式可以在罌粟育種中,培育出不含嗎啡,以及提高其他生物鹼含量的品種。丹參 (Salvia miltiorrhiza)為傳統中藥,用於祛瘀止痛、活血化瘀和養血安神等作用。近年來用來治療心血管疾病、缺血性中風、肝炎和慢性腎功能衰竭等疾病。將生合成丹參酮 (tanshinone)的基因committed diterpene synthase (SmCPS1)以CRISPR/Cas9 方法剔除,之後以LC-qTOF-MS 和 Q-TRAP-LC-MS/MS分析該丹參突變株,cryptotanshinone (隱丹參酮)、tanshinone IIA (丹參酮 IIA) 和 tanshinone I (丹參酮 I)幾乎偵測不到,但其他酚酸代謝物如salvianolic acid B (丹酚酸B)生合成不受任何的影響。利用基因編輯方式,將來可以使得丹參有效的次級代謝物可以大量的生合成出來 (Li et al., 2017)。另外,將丹參中的生合成水溶性酚酸中的關鍵酵素基因rosmarinic acid synthase (SmRAS)剔除後,會使得rosmarinic acid (迷迭香酸) 和lithospermic acid B (紫草酸B)明顯的生合成量下降,而其前驅物3,4-dihydroxyphenyllactic acid 含量則顯著的增加 (Zhou et al., 2018)。
未來展望
臺灣觀賞作物產業近年來快速的發展,其中又以蘭花為重要外銷商品,因此對於觀賞作物育種及栽培技術和特性如外觀、香味、花色、抗病性和生理特性等研究需求也日益殷切,若能透過分子標記及基因工程方式來加速臺灣新穎花卉育種產生多樣式的花卉外觀、新穎的顏色和改善香氣等。國外消費者對於基因轉殖所育成上市的藍色玫瑰和康乃馨,具高接受度。臺灣在花卉育種方面除了可以發展分子標記輔助育種外,也可以利用基因轉殖技術來開發新型的花卉品種。此外,新發展出的基因編輯技術如 CRISPR/Cas9,可精準的編輯作物基因組,操作簡單、便宜、快速,因此被視為作物改良的利器。美國食品與藥物管理局宣布不會將基因編輯作物(GE crops)視為轉基因作物(GM crops)。蘭花為高經濟價值花卉,其中又以蝴蝶蘭為臺灣蘭花產業中產值最高,而育種出新穎的蝴蝶蘭品種平均要花費 3~4 年的時間,如果能夠透過基因編輯技術改變如蝴蝶蘭的多梗性、多花、抗病、花色、花型、開花時間及植株構型(plant architecture),可以加速蝴蝶蘭的育種。


參考文獻
Alagoz, Y., Gurkok, T., Zhang, B. and Unver, T. (2016) Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in opium poppy using CRISPR-Cas 9 genome editing technology. Sci. Rep. 6, 1-9.
Bolotin, A., B. Quinquis, A. Sorokin, and S. D. Ehrlich. (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551-2561.
Chen, F., Song, Y., Li, X., Chen, J., Mo, L., Zhang, X., Lin, Z. and Zhang, L. (2019) Genome sequences of horticultural plants: past, present, and future. Hortic. Res. 6.
De-Eknamkul, W. and Zenk, M.H. (1992) Purification and properties of 1, 2-dehydroreticuline reductase from Papaver somniferum seedlings. Phytochemistry 31, 813-821.
Deveau, H., J. E. Garneau, and S. Moineau. (2010) CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 64, 475-493.
Goodstein, D.M., Shu, S., Howson, R., Neupane, R., Hayes, R.D., Fazo, J., Mitros, T., Dirks, W., Hellsten, U. and Putnam, N. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178-D1186.
Dey, A., Bhattacharya, R., Mukherjee, A. and Pandey, D.K. (2017) Natural products against Alzheimer’s disease: Pharmaco-therapeutics and biotechnological interventions. Biotechnol. Adv. 35, 178-216.
Dey, A., Nandy, S., Mukherjee, A. and Modak, B.K. (2021) Sustainable utilization of medicinal plants and conservation strategies practiced by the aboriginals of Purulia district, India: a case study on therapeutics used against some tropical otorhinolaryngologic and ophthalmic disorders. Environ. Dev. Susta. 23, 5576-5613.
Duprat, A., Caranta, C., Revers, F., Menand, B., Browning, K.S. and Robaglia, C. (2002) The Arabidopsis eukaryotic initiation factor (iso) 4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32, 927-934.
Fairbairn, J. and Wassel, G. (1964) The alkaloids of Papaver somniferum L.-I.: Evidence for a rapid turnover of the major alkaloids. Phytochemistry 3, 253-258.
Feng, J., C. Dai, H. Luo, Y. Han, Z. Liu, and C. Kang. (2018) Reporter gene expression reveals precise auxin synthesis sites during fruit and root development in wild strawberry. J. Exp. Bot. 70, 563-574.
Feng, Z., Y. Mao, N. Xu, B. Zhang, P. Wei, D.-L. Yang and all. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 111, 4632-4637.
Gerdemann, C., Eicken, C. and Krebs, B. (2002) The crystal structure of catechol oxidase: new insight into the function of type-3 copper proteins. Acc. Chem. Res. 35, 183-191.
Gómez, P., Rodríguez-Hernández, A., Moury, B. and Aranda, M. (2009) Genetic resistance for the sustainable control of plant virus diseases: breeding, mechanisms and durability. Eur. J. Plant. Pathol. 125, 1-22.
González, M.N., Massa, G.A., Andersson, M., Turesson, H., Olsson, N., Fält, A.-S., Storani, L., Décima Oneto, C.A., Hofvander, P. and Feingold, S.E. (2020) Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant Sci. 10, 1649.
Hu, Y., J. Zhang, H. Jia, D. Sosso, T. Li, W. B. Frommer and all. (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 111, E521-E529.
Jackson, R.J., Hellen, C.U. and Pestova, T.V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113-127.
Jinek, M., K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna, and E. Charpentier. (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821.
Kaur, N., A. Alok, Shivani, N. Kaur, P. Pandey, P. Awasthi and all. (2018) CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genomics 18, 89-99.
Klimek-Chodacka, M., T. Oleszkiewicz, L. G. Lowder, Y. Qi, and R. Baranski. (2018) Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 37, 575-586.
Lellis, A.D., Kasschau, K.D., Whitham, S.A. and Carrington, J.C. (2002) Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF (iso) 4E during potyvirus infection. Curr. Biol. 12, 1046-1051.
Levy, S. E., and R. M. Myers. (2016) Advancements in next-generation sequencing. Annu. Rev. Genomics Hum. Genet. 17, 95-115.
Li, B., Cui, G., Shen, G., Zhan, Z., Huang, L., Chen, J. and Qi, X. (2017) Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza. Sci. Rep. 7, 1-9.
Li, J.F., J. E. Norville, J. Aach, M. McCormack, D. Zhang, J. Bush and all. (2013) Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688.
Li, R., Li, R., Li, X., Fu, D., Zhu, B., Tian, H., Luo, Y. and Zhu, H. (2018) Multiplexed CRISPR/Cas9‐mediated metabolic engineering of γ‐aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. 16, 415-427.
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., Luo, Y. and Zhu, H. (2018) Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-Mediated Multiplex Genome Editing. Front. Plant Sci. 9, 559.
Maioli, A., Gianoglio, S., Moglia, A., Acquadro, A., Valentino, D., Milani, A.M., Prohens, J., Orzaez, D., Granell, A. and Lanteri, S. (2020) Simultaneous CRISPR/Cas9 Editing of Three PPO Genes Reduces Fruit Flesh Browning in Solanum melongena L. Front. Plant Sci. 11, 607161.
Marks, R.A., Hotaling, S., Frandsen, P.B. and VanBuren, R. (2021) Representation and participation across 20 years of plant genome sequencing. Nature plants 7, 1571-1578.
Mojica, F. J. M., C. s. Díez-Villaseñor, J. García-Martínez, and E. Soria. (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol.
Morishige, T., Tsujita, T., Yamada, Y. and Sato, F. (2000) Molecular characterization of the S-adenosyl-L-methionine: 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 275, 23398-23405.
Nakajima, I., Y. Ban, A. Azuma, N. Onoue, T. Moriguchi, T. Yamamoto and all. (2017) CRISPR/Cas9-mediated targeted mutagenesis in grape. PLOS One 12, 0177966.
Nicaise, V., German-Retana, S., Sanjuán, R., Dubrana, M.-P., Mazier, M., Maisonneuve, B., Candresse, T., Caranta, C. and LeGall, O. (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol. 132, 1272-1282.
Nishihara, M., A. Higuchi, A. Watanabe, and K. Tasaki. 2018. Application of the CRISPR/Cas9 system for modification of flower color in Torenia fournieri. BMC Plant Biol. 18, 331.
Nishitani, C., N. Hirai, S. Komori, M. Wada, K. Okada, K. Osakabe and all. (2016) Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6, 31481.
Nonaka, S., Arai, C., Takayama, M., Matsukura, C., Ezura, H. (2017) Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci Rep. 7, 7057.
Peng, A., S. Chen, T. Lei, L. Xu, Y. He, L. Wu and all. (2017) Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. 15, 1509-1519.
Pourcel, C., G. Salvignol, and G. Vergnaud. (2005) CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653-663.
Ruffel, S., Gallois, J.-L., Moury, B., Robaglia, C., Palloix, A. and Caranta, C. (2006) Simultaneous mutations in translation initiation factors eIF4E and eIF (iso) 4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87, 2089-2098.
Sanfaçon, H. (2015) Plant translation factors and virus resistance. Viruses 7, 3392-3419.
Shan, Q., Y. Wang, J. Li, Y. Zhang, K. Chen, Z. Liang and all. (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686-688.
Singh, P. and Ali, S.A. (2021) Impact of CRISPR-Cas9-Based Genome Engineering in Farm Animals. Vet Sci. 8:122.
Tian, S., L. Jiang, Q. Gao, J. Zhang, M. Zong, H. Zhang and all. (2017) Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 36, 399-406.
Tashkandi, M., Ali, Z., Aljedaani, F., Shami, A. and Mahfouz, M.M. (2018) Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 13, e1525996.
Tripathi, J. N., V. O. Ntui, M. Ron, S. K. Muiruri, A. Britt, and L. Tripathi. (2019) CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2, 46.
Valva, V.L., Sabato, S. and Gigliano, G.S. (1985) Morphology and alkaloid chemistry of Papaver setigerum DC.(Papaveraceae). Taxon 34, 191-196
Wan, D.-Y., Guo, Y., Cheng, Y., Hu, Y., Xiao, S., Wang, Y. and Wen, Y.-Q. (2020) CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic. Res. 7.
Wang, A. and Krishnaswamy, S. (2012) Eukaryotic translation initiation factor 4E‐mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795-803.
Wang, Z., S. Wang, D. Li, Q. Zhang, L. Li, C. Zhong and all. (2018) Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. 16, 1424-1433.
Watanabe, K., A. Kobayashi, M. Endo, K. Sage-Ono, S. Toki, and M. Ono. (2017) CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci. Rep. 7, 10028.
Wiedenheft, B., S. H. Sternberg, and J. A. Doudna. (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331-338.
Wilson, F. M., K. Harrison, A. D. Armitage, A. J. Simkin, and R. J. Harrison. (2019) CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15, 45.
Zhang, Y., Z. Liang, Y. Zong, Y. Wang, J. Liu, K. Chen and all. (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617.
Zhou, Z., Tan, H., Li, Q., Chen, J., Gao, S., Wang, Y., Chen, W. and Zhang, L. (2018) CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry 148, 63-70.
作者簡介

專長為蘭花生物技術、植物組織培養、精準育種技術

